A graph consists of a set of vertices and edges. Each edge connects two vertices. In a undirected graph, edges have no direction: two vertices are either connected by an edge or they are not. In a directed graph, each edge has a direction: it points from one vertex to another. A graph is dense if it has many edges relative to its number of vertices, and sparse if it has few such edges. Some graphs are weighted, in which case every edge has a number called its weight.
Graphs are fundamental in computer science. Many problems can be expressed in terms of graphs, and we can answer many interesting questions using graph algorithms.
Often we will assume that each vertex in a graph has a unique id. Sometimes ids are integers; in a graph with V vertices, typically these integers will range from 0 through (V - 1). Here is one such undirected graph:
Alternatively, we may sometimes assume that vertex ids are strings, or belong to some other data type.
How may we store a graph with V vertices in
memory? As one possibility, we can use adjacency-matrix
representation. Assuming that each vertex has an integer id, the
graph will be stored as a two-dimensional array
g of booleans, with dimensions V
x V. In Python, g will be a list of lists. In this representation,
g[i][j] is true if and only there is an edge from vertex
i to vertex j.
If the graph is undirected, then the adjacency
matrix will be symmetric: g[i][j] = g[j][i]
for all i and j.
Here is an adjacency-matrix representation of the undirected graph above. Notice that it is symmetric:
graph = [
[False, True, True, False, False, True, True],
[True, False, False, False, False, False, False],
[True, False, False, False, False, False, False],
[False, False, False, False, True, True, False],
[False, False, False, True, False, True, True],
[True, False, False, True, True, False, False],
[True, False, False, False, True, False, False]
]Also notice that all the entries along the main diagonal are False. That's because the graph has no self-edges, i.e. edges from a vertex to itself.
Alternatively,
we can use adjacency-list representation, in which for
each vertex we store an adjacency list, i.e. a list of its
adjacent vertices. Assuming that vertex ids are integers, then
in Python this representation will generally
take the form of a list
of lists
of integers. For a graph g, g[i]
is the adjacency list of i, i.e. a list of ids of vertices adjacent
to i. (If the graph is directed, then more specifically g[i] is a
list of vertices to which there are edges leading from i.)
When we store an undirected graph in adjacency-list representation, then if there is an edge from u to v we store u in v’s adjacency list and also store v in u’s adjacency list.
Here is an adjacency-list representation of the undirected graph above:
graph = [
[1, 2, 5, 6], # neighbors of 0
[0], # neighbors of 1
[0], # …
[4, 5],
[3, 5, 6],
[0, 3, 4],
[0, 4]
]Now suppose that vertex ids are not integers – for example, they may be strings, as in this graph:
Then in Python the adjacency-list representation will be a dictionary of lists. For example, the graph above has this representation:
graph = {
'c': ['f', 'q', 'x'],
'f': ['c', 'p', 'q', 'w'],
'p': ['f'],
'w': ['f', 'q', 'x'],
'x': ['c', 'w']
}Suppose that we have a graph with integer ids. Which representation should we use? Adjacency-matrix representation allows us to immediately tell whether two vertices are connected, but it may take O(V) time to enumerate a given vertex’s edges. On the other hand, adjacency-list representation is more compact for sparse graphs. With this representation, if a vertex has e edges then we can enumerate them in time O(e), but determining whether two vertices are connected may take time O(e), where e is the number of edges of either vertex. Thus, the running time of some algorithms may differ depending on which representation we use.
In this course we will usually represent graphs using adjacency-list representation.
In this course we will study two fundamental algorithms that can search a directed or undirected graph, i.e. explore it by visiting its nodes in some order.
The first of these algorithms is depth-first search, which is something like exploring a maze by walking through it. Informally, it works like this. We start at any vertex, and begin walking along edges. As we do so, we remember all vertices that we have ever visited. We will never follow an edge that leads to an already visited vertex. Each time we hit a dead end, we walk back to the previous vertex, and try the next edge from that vertex (as always, ignoring edges that lead to visited vertices). If we have already explored all edges from that vertex, we walk back to the vertex before that, and so on.
For example, consider this undirected graph that we saw above:
Suppose that we perform a depth-first search of this graph, starting with vertex 0. We might first walk to vertex 1, which is a dead end. The we backtrack to 0 and walk to vertex 2, which is also a dead end. Now we backtrack to 0 again and walk to vertex 5. From there we might proceed to vertex 3, then 4. From 4 we cannot walk to 5, since we have been there before. So we walk to vertex 6. From 6 we can't proceed to 0, since we have been there before. So we backtrack to 4, then back to 3, 5, and then 0. The depth-first search is complete, since there are no more edges that lead to unvisited vertices.
As a depth-first search runs, we may imagine that at each moment every one of its vertices is in three possible states:
A vertex is undiscovered if we have not yet seen it.
A vertex V is on the frontier if we have seen it, and are still exploring some part of the graph that we reached through an edge leading from V. In other words, we will backtrack to V at some future time.
A vertex is explored if we have left it because we have fully explored all possible paths leading from it, so we will not backtrack to it again.
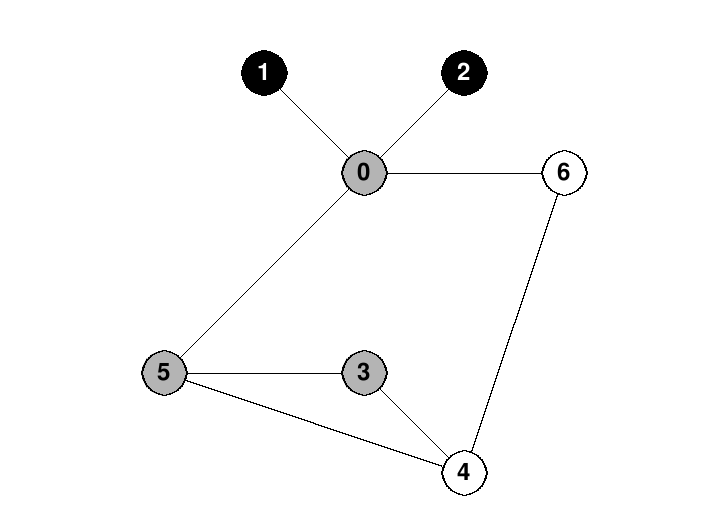
Often when we draw pictures of a depth-first search (and also of some other graph algorithms), we draw undiscovered vertices in white, frontier vertices in gray and explored vertices in black.
For example, here is a picture of a depth-first search in progress on the graph above. The search started at vertex 0, went to vertex 1, then backtracked back to 0. Then it went to vertex 2 and backtracked back to 0 again. Then it proceeded to vertex 5, and has just arrived at vertex 3:
Here's a larger graph, with vertices representing countries in Europe. The graph contains edges between countries that either border each other or are reachable from each other by a direct ferry (in which case the edges are depicted as dashed lines):
Here's a picture of a depth-first search in progress on this graph. The search started with Austria, and has now reached Italy:
It is easy to implement a depth-first search using a recursive function. However, we must avoid walking through a cycle in an infinite loop. To accomplish this, as described above, when we first visit a vertex we mark it as visited. Whenever we follow an edge, if it leads to a visited vertex then we ignore it.
If a graph has vertices numbered from 0 to (V – 1), we may efficiently represent the visited set using an array of V boolean values. Each value in this array is True if the corresponding vertex has been visited.
Here is Python code that implements a depth-first search using a nested recursive function. It takes a graph g in adjacency-list representation, plus a vertex id 'start' where the search should begin. It assumes that vertices are numbered from 0 to (V – 1), where V is the total number of vertices.
# recursive depth-first search over graph with integer ids
def dfs(g, start):
visited = [False] * len(g)
def visit(v):
print('visiting', v)
visited[v] = True
for w in g[v]: # for all neighbors of vertex v
if not visited[w]:
visit(w)
visit(start)This function merely prints out the visited nodes. But we can modify it to perform useful tasks. For example, to determine whether a graph is connected, we can run a depth-first search and then check whether the search visited every node. As you will see in more advanced courses, a depth-first search is also a useful building block for other graph algorithms: determining whether a graph is cyclic, topological sorting, discovering strongly connnected components and so on.
This depth-first search does a constant amount of work for each vertex (making a recursive call) and for each edge (following the edge and checking whether the vertex it points to has been visited). So it runs in time O(V + E), where V and E are the numbers of vertices and edges in the graph.
As another example, let's run a depth-first search on the undirected graph of Europe that we saw above. Here is a file 'europe' containing data that we can use to build the graph. The file begins like this:
austria: germany czech slovakia hungary slovenia italy belgium: netherlands germany luxembourg france bulgaria: romania greece …
And here is a function that can read this file to build an adjacency-list representation in memory:
# read graph in adjacency list representation
def read_graph(file):
g = {}
def add(v, w):
if v not in g:
g[v] = []
g[v].append(w)
with open(file) as f:
for line in f:
source, dest = line.split(':')
if source not in g:
g[source] = []
for c in dest.split():
add(source, c)
add(c, source)
return gHere is a function that will perform the depth-first search. Since vertex ids are strings, we can't represent the visited set using an array of booleans as we did above; instead, we use a Python set.
# recursive depth-first search over graph with arbitrary ids
def dfs(g, start):
visited = set()
def visit(v, depth):
print(' ' * depth + v)
visited.add(v)
for w in g[v]:
if w not in visited:
visit(w, depth + 1)
visit(start, 0)The recursive helper function visit() uses an extra parameter 'depth' to keep track of the search depth. When it prints the id of each visited vertex, it indents the output by 2 spaces for each indentation level. Let's run the search:
>>> europe = read_graph('europe')
>>> dfs(europe, 'ireland')
ireland
uk
france
belgium
netherlands
germany
austria
czech
poland
lithuania
latvia
estonia
finland
sweden
denmark
slovakia
hungary
croatia
slovenia
italy
greece
bulgaria
romania
malta
luxembourg
spain
portugal
>>> The indentation reveals the recursive call structure. We see that from the start vertex 'ireland' we recurse all the way down to 'denmark', then backtrack to 'poland' and begin exploring through 'slovakia', and so on.
Even with these changes to work with arbitary vertex ids, our depth-first search code still runs in O(V + E), since dictionary and set operations in Python (e.g. looking up an element in a dictionary, or checking whether an element is present in a set) run in O(1) on average.
Note that Python has a recursion limit of 1000 calls, so it would be unwise to run a recursive depth-first search in Python on a graph with more then 1000 vertices. As an alternative, it is also possible to implement a depth-first search non-recursively. We'll see how in the next lecture.
In the last lecture we learned about graph representations and depth-first search. We will now look at breadth-first search, which is another fundamental graph algorithm.
Starting from some vertex V, a breadth-first search first visits vertices adjacent to V, i.e. vertices of distance 1 from V. It then visits vertices of distance 2, and so on. In this way it can determine the shortest distance from V to every other vertex in the graph.
We can implement a breadth-first search using a queue, i.e. a FIFO (first in first out) data structure. Also, just as with a depth-first search, we will need a visited set to avoid walking in circles as we search.
We begin by adding the start vertex to the queue and marking it as visited. In a loop, we repeatedly remove vertices from the queue. Each time we remove a vertex, we mark all of its adjacent unvisited vertices as visited and add them to the queue. The algorithm terminates once the queue is empty, at which point we will have visited all reachable vertices.
For example, consider the graph of Europe that we saw in the previous lecture:
Suppose that we begin a breadth-first search at 'austria'. Initially the queue contains only that vertex:
Q = austria
We dequeue 'austria' and enqueue its neighbors (and add them to the visited set):
Q = germany czech hungary slovakia slovenia italy
Next, we dequeue 'germany' and add its neighbors to the queue. However we do not add 'austria' or 'czech', which have already been visited:
Q = czech hungary slovakia slovenia italy belgium netherlands luxembourg denmark poland france
Now we dequeue 'czech'. Its neighbors are 'austria', 'germany', 'poland' and 'slovakia' but they are all visited, so there is nothing to add:
Q = hungary slovakia slovenia italy belgium netherlands luxembourg denmark poland france
Now we dequeue 'hungary'. It has two unvisited neighbors 'romania' and 'croatia', so we enqueue them:
Q = slovakia slovenia italy belgium netherlands luxembourg denmark poland france romania croatia
Next, we dequeue 'slovakia' and 'slovenia'. They have no unvisited neighbors:
Q = italy belgium netherlands luxembourg denmark poland france romania croatia
Just as with a depth-first search, we may imagine that at any time during a breadth-first search, each vertex is either undiscovered (white), on the frontier (gray) or explored (black). The queue represents the frontier, i.e. the gray vertices. So the picture now looks like this:
The gray vertices are in the queue; the black vertices have already been dequeued; the white vertices have not yet entered the queue.
As the algorithm runs, all vertices in the queue are at approximately the same distance from the start vertex. To be more precise, at every moment in time there is some value d such that all vertices in the queue are at distance d or (d + 1) from the start vertex. (At this particular moment in this example, all the vertices in the queue have distance either 1 or 2 from the start vertex 'austria'.)
To implement a breadth-first search in Python, we need a queue implementation. As we learned in a previous lecture, we could implement an efficient queue using a linked list. However, Python's library contains a type deque that we can use instead, which is a bit more convenient. A deque is a double-ended queue: at any time we can append or prepend an item, or remove an item from the front or back of the queue. All of these operations are efficient, i.e. run in O(1) on average. In our implementation, we will call the append() method of the deque class to add an element to the queue, and will call popleft() to remove an item. (Alternatively, we could call appendleft() and pop(), in which case our items would travel through the deque in the opposite direction.)
Here's an implementation of breadth-first search for a graph with integer ids. The visited set is an array of booleans:
from collections import deque
# breadth-first search over graph with integer ids
def bfs(graph, start):
visited = [False] * len(graph)
visited[start] = True
q = deque()
q.append(start)
while len(q) > 0:
v = q.popleft()
print('visiting', v)
for w in graph[v]:
if not visited[w]:
visited[w] = True
q.append(w)Notice that we mark vertices as visited when we add them to the queue, not when we remove them. This ensures that our algorithm will not add the same vertex to the queue more than once.
Like a depth-first search, this breadth-first search function does a constant amount of work for each vertex and edge, so it also runs in time O(V + E).
Let's modify our breadth-first search code so that it can run on our graph of Europe. Since vertex ids are strings, we will store the visited set using a Python set, not an array of booleans. Also, as we search we will compute the distance from the start vertex to every other vertex that we reach. To achieve this, each queue element will be a pair (v, d), where v is a vertex id and d is the distance from the start vertex to v. Finally, when we print out each vertex we will indent it by its distance from the start. Here is the code:
from collections import deque
# breadth-first search over graph with arbitrary ids
def bfs(g, start):
visited = { start }
q = deque()
q.append((start, 0))
while len(q) > 0:
v, dist = q.popleft()
print(' ' * dist + v)
for w in g[v]:
if w not in visited:
q.append((w, dist + 1))
visited.add(w)Let's run it:
>>> bfs(europe, 'ireland')
ireland
uk
france
belgium
luxembourg
germany
italy
spain
netherlands
austria
czech
denmark
poland
greece
malta
slovenia
portugal
slovakia
hungary
sweden
lithuania
bulgaria
croatia
romania
finland
latvia
estonia
>>> We can easily see that the breadth-first search discovers vertices in increasing order of distance from the start vertex.
As it runs, a breadth-first search implicitly generates a breadth-first tree, in which each vertex points to all vertices that are first discovered from it as we search. Here is a breadth-first tree generated by searching our graph of European countries, starting with Austria:
In this picture, the dotted lines are not part of the breadth-first tree. They are edges which were not followed during the search, since they pointed to vertices that were already in the visited set.
The breadth-first tree contains a shortest path from the starting vertex ('austria') to every other vertex in the graph.
Let's now reverse all edges in the breadth-first tree. The result looks like this:
In this picture, each vertex points to its predecessor, namely the vertex from which we discovered it during the breadth-first search. Starting with any vertex v, we can follow arrows to reach the starting vertex. As we do so, we will be walking along a shortest path from v to the starting vertex.
We can modify our breadth-first search code so that as it runs, it remembers the predecessor of each vertex that it discovers. Then once we reach a goal vertex we can retrace a path back to the starting vertex. If we reverse this path, we will have a path from the start to the goal. Here is the updated code:
from collections import deque
# Return a list of vertices along a shortest path from the given start vertex
# to the given goal. If there is no such path, return None.
def bfs_path(graph, start, goal):
visited = { start }
pred = {}
q = deque()
q.append(start)
pred[start] = None # start vertex has no predecessor
while len(q) > 0:
v = q.popleft()
if v == goal:
path = []
while True:
path.append(v)
if pred[v] == None:
break
v = pred[v]
path.reverse()
return path
for w in graph[v]:
if w not in visited:
q.append(w)
visited.add(w)
pred[w] = v # remember w's predecessor
return None # no pathLet's take our breadth-first search code from above, and modify it so that it uses a stack instead of a queue. We can use an ordinary Python list as a stack: we'll call append() to push and pop() to pop. Both of these operations will run in O(1) on average.
from collections import deque
def search(g, start):
visited = { start }
q = []
q.append(start)
while len(q) > 0:
v = q.pop()
print(f'visiting {v}')
for w in g[v]:
if w not in visited:
q.append(w)
visited.add(w)Let's run this code on our Europe graph:
>>> search(graph, 'austria') visiting austria visiting italy visiting malta visiting greece visiting bulgaria visiting romania ...
The code performs a depth-first search! This shows how we can perform a depth-first search non-recursively. This is useful, since Python (and many other languages) allow only a limited depth of recursive calls.
Now, this depth-first search code will not visit the vertices in the same order as the recursive depth-first search that we wrote before. That's because it processes a vertex's neighbors in a slightly different way than the recursive version does. In particular, this version marks all neighboring vertices as visited before it explores paths through any of them. As a somewhat challenging exercise, you could attempt to modify this non-recursive code so that it visits vertices in the exact same order as the recursive version.